Posts by Natalia
|
1)
Message boards :
Number crunching :
New Work
(Message 2463)
Posted 9 Jun 2025 by  Natalia Natalia
Post: Hello! We are preparing a new target, but it takes longer than we expected. Also, the team is working on the results previously obtained in SiDock@home. |
|
2)
Message boards :
News :
Temporarily work unavailabilities
(Message 2443)
Posted 4 Apr 2025 by  Natalia Natalia
Post: We hope there will be new tasks available in April or May. |
|
3)
Message boards :
Number crunching :
what is corona_Sprot_delta_v1?
(Message 2422)
Posted 24 Feb 2025 by  Natalia Natalia
Post: Hello! The target corona_Sprot_delta_v1 is related to human coronavirus SARS-CoV-2. It is the RBD (receptor-binding domain) Sprot domain surface interacting with ACE2. |
|
4)
Message boards :
Number crunching :
How Big is Target #23: Ebola GP1?
(Message 2421)
Posted 24 Feb 2025 by  Natalia Natalia
Post: Hello! I apologize for the late response. For each target, including Ebola GP1, we perform a virtual screening of a library containing 982,401,500 molecules. The library is divided into units of 500 molecules. Each unit, approximately 240 KB in size, serves as input for one BOINC workunit. |
|
5)
Message boards :
News :
Changes in task scheduling and monitoring mechanism
(Message 2304)
Posted 22 Jul 2024 by  Natalia Natalia
Post: Hello! BOINC client displays the deadline in your local time, as the computer it is running on. A task which misses the deadline still can be useful (and earn BOINC credit) for several days. |
|
6)
Message boards :
News :
Target # 23: Ebola GP1
(Message 2284)
Posted 11 Jul 2024 by  Natalia Natalia
Post: Dear participants, as target # 22 is almost finished, we are glad to introduce the next target. Most of you have voted for Ebolavirus glycoprotein (GP) (174 out of 425 votes, wow!) The Ebola virus is a highly virulent pathogen responsible for causing Ebola hemorrhagic fever, a severe and often fatal disease. A key factor in the virus's ability to infect host cells and cause disease is its surface glycoprotein (GP), making it an attractive target for antiviral drug development. The Ebola GP is a trimeric protein composed of two subunits per monomer: GP1, responsible for receptor binding, and GP2, which mediates fusion between the viral and host cell membranes. Initially synthesized as a precursor protein, the GP is cleaved by host proteases (furin, cathepsin) into its functional subunits, a process essential for its role in mediating viral entry. The GP facilitates the virus's attachment to the host cell surface, followed by conformational changes that enable membrane fusion, allowing the virus to enter the host cell (to the host endosomal Niemann-Pick C1 (NPC1) receptor or via direct membrane binding; Vaknin et al.; ACS Infect. Dis. 2024, 10, 5, 1590–1601). Targeting the GP for drug development is advantageous due to its essential role in viral infection, its highly conserved structure among different Ebola virus strains, and the availability of specific binding cavities that can accommodate small-molecule inhibitors. Structural studies using techniques such as X-ray crystallography have identified these binding cavities and elucidated the GP's conformation in both its free and inhibited states. These insights enable the design of drugs that can specifically bind to and inhibit the GP by stabilizing it in its pre-fusion conformation or interfering with its cleavage, thereby preventing the necessary conformational changes for membrane fusion. We will employ high-resolution structures to conduct virtual screening experiments coupled to molecular dynamics simulations to ultimately identify potential GP inhibitors/modulators. Promising compounds identified through these computational methods will hopefully undergo further validation using biochemical assays, pseudovirus entry assays, and structural analyses to confirm their inhibitory activity. Targeting the GP offers specificity, as it minimizes off-target effects on host cells and reduces the likelihood of resistance development. Moreover, due to the conserved nature of the GP, drugs targeting it could be effective against multiple Ebola virus strains and variants. We hope that our computations will contribute to the fight against Ebola! |
|
7)
Message boards :
News :
Target # 22: corona_RdRp_v2
(Message 2282)
Posted 10 Jul 2024 by  Natalia Natalia
Post: We already have a new target for fighting the Ebola virus :) |
|
8)
Message boards :
Number crunching :
Just wanted to say "Thank you"
(Message 2267)
Posted 22 May 2024 by  Natalia Natalia
Post: Thank you for the support! Virtual screening produces lots of computational tasks, and we hope that our results will lead to new knowledge and progress in fighting the diseases. |
|
9)
Message boards :
Cafe :
BOINC Competition for High School Students - Computation Moonshot
(Message 2204)
Posted 13 Feb 2024 by  Natalia Natalia
Post: Great news, let this competition attract new crunchers and raise public interest to scientific computing. |
|
10)
Message boards :
News :
Vote for the next target!
(Message 2176)
Posted 26 Jan 2024 by  Natalia Natalia
Post: Dear all, we have obtained results for different targets of SARS-CoV-2. The computations continue, and we want to ask your opinion on the next target. You can vote for one of them until February 5th, 2024.
|
|
11)
Message boards :
News :
Project status: December 2023
(Message 2154)
Posted 26 Dec 2023 by  Natalia Natalia
Post: Dear SiDock@home participants, We've successfully achieved 21 milestones in our ongoing drug discovery initiative, and this strong, open, and community-supported drug discovery project is going on. Our research has been routine lately: virtual screening on the same library for a pleiade of corona-related targets. These efforts are however crucial to our research progression. Currently, we're in the process of drafting publications for two of our completed objectives (3CLpro and PLpro) and are setting the stage for upcoming drug targets (here we are also planning a pool where You the participants, will help us decide on the upcoming target work). Last but not least, we thank everyone who has donated cryptocurrency or money. The donations sum up to 639 Euro and 12,140 Gridcoin now. We plan to use them for purchase of compounds and in-vitro screening. As always, we are grateful to all of you for your computational contributions and discussions! We look forward to the future work on SiDock@home. Merry Christmas to all and all the best to All! With best wishes, Natalia, Marko, Črtomir and hoarfrost 
|
|
12)
Message boards :
News :
BOINC merchandise by the Science Commons Initiative
(Message 2005)
Posted 23 Feb 2023 by  Natalia Natalia
Post: Dear participants, There is a wonderful new beginning started by the Science Commons Initiative. They launched a web store selling BOINC-related merchandise. SiDock@home is already on board :) We support this idea and hope it will help more people to know about BOINC and get interested in BOINC projects! It will also contribute to the development of BOINC as 50% of the profits will go to the BOINC Development Fund (100% in special cases). Another 50% of the profit will go to the project as a donation. With best wishes, team of SiDock@home |
|
13)
Message boards :
News :
BOINC Census 2022, be counted!
(Message 1985)
Posted 10 Feb 2023 by  Natalia Natalia
Post: An update from the Science Commons Initiative: BOINC Census Released! |
|
14)
Message boards :
Science :
Other uses of docking than viruses?
(Message 1943)
Posted 24 Jan 2023 by  Natalia Natalia
Post: Hello! GPU version of CmDock is under development. The latest CPU version is CmDock v0.2.0, December 2022. |
|
15)
Message boards :
News :
Merry Christmas!
(Message 1776)
Posted 24 Dec 2022 by  Natalia Natalia
Post: Dear participants, We wish you a merry Christmas and a happy New Year! With your help, we have performed a lot of computer modeling. We are going to continue, and believe the results will lead to advances in drug development! For now, we have a fresh new release of CmDock and will employ it starting from the next target. Here is an illustration of the modeling performed in the project, visualized with PyMOL. You can see a large molecule (this is the target, TMPRSS2 protein) with a molecule many times smaller. The small molecules are ligands, each of them can interact with the target in its own way, with its own binding energy. The more precisely the ligand binds to the target, the higher the chance that it will be able to change the functioning of the large molecule. For example, the TMPRSS2 enzyme shown in the image is necessary for a chemical reaction allowing the coronavirus to enter the body cells and, accordingly, a substance that can suppress the normal operation of this enzyme can make it difficult or block the spread of the coronavirus in it. During the modeling performed on your computers, various ligands from the library are tested, and the best ones are selected to process on next stages of drug development. All the best, Natalia, Marko, hoarfrost, Črtomir 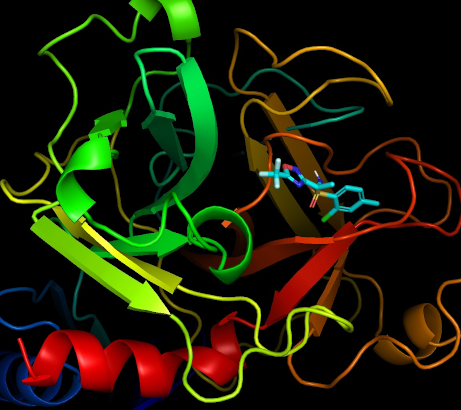
|
|
16)
Message boards :
News :
BOINC Census 2022, be counted!
(Message 1761)
Posted 17 Nov 2022 by  Natalia Natalia
Post: The BOINC Census is a yearly effort aimed at finding out who uses BOINC, what they like about it, and what changes they want to see in the future. Filling out the census is a great way to support this project, the wider BOINC community, and have your input heard for future bug bounties. The census is sponsored by the Science Commons Initiative, and the results will be published in an aggregate, anonymized fashion at the completion of the census. Please take 5 minutes to fill out the census at https://app.fillout.com/flow/esDGuRswWfus |
|
17)
Message boards :
News :
Target 21: corona_TMPRSS2_v1
(Message 1760)
Posted 17 Nov 2022 by  Natalia Natalia
Post: Dear Participants, TMPRSS2 is a type II transmembrane protease with broad expression in epithelial cells of the respiratory and gastrointestinal human tracts. It is a cofactor in SARS-CoV-2 entry, and primes viral proteins. Namely, TMPRSS2 cleaves the viral Spro to expose the fusion peptide for cell entry. We are therefore examining TMPRSS2 as a promising strategy to block viral infection in a host-directed therapeutic and/or prophylactic manner. This work is also a a part of larger viral-host protease inhibition study campaign. With best wishes, Team SiDock@home |
|
18)
Questions and Answers :
Windows :
Will not queue more than about 4 work units per core despite set for 10 days of work
(Message 1716)
Posted 24 Aug 2022 by  Natalia Natalia
Post: At the beginning, we followed the list of targets (groups of work units) that was published in the presentation. Today, new targets are selected following the intermediate computational results and new scientific data. For instance, the target PLpro was investigated 4 times instead of 2 planned initially. This is why it is difficult to publish a planned list in advance. |
|
19)
Questions and Answers :
Web site :
Email notification broken
(Message 1696)
Posted 12 Aug 2022 by  Natalia Natalia
Post: Email notifications should be fixed now, please check. |
|
20)
Questions and Answers :
Web site :
Update Cross-Project ID
(Message 1652)
Posted 21 Jun 2022 by  Natalia Natalia
Post: Hello, as I gather, only you can fix it at your side. Following this post, you can either (1) ensure your email addresses match across all the projects you signed up to, connect all your projects to one host and wait 'up to 14 days' for the CPIDs to settle. Or (2) follow the steps listed in the post to fix it immediately. Please tell us if you managed to fix it. |
Next 20

©2025 SiDock@home Team