News archive
Temporarily no donations to SiDock@home in cryptocurrency
Dear participants,
we temporarily stop accepting donations to SiDock@home in cryptocurrency. If you have automated donations to SiDock@home in cryptocurrency, please disable them. Our previous GRC/BTC/ETH addresses will be unavailable.
With best wishes,
Team of SiDock@home
9 Oct 2025, 9:25:07 UTC
· Discuss
Target # 23: Ebola GP1
Dear participants,
as target # 22 is almost finished, we are glad to introduce the next target. Most of you have voted for Ebolavirus glycoprotein (GP) (174 out of 425 votes, wow!)
The Ebola virus is a highly virulent pathogen responsible for causing Ebola hemorrhagic fever, a severe and often fatal disease. A key factor in the virus's ability to infect host cells and cause disease is its surface glycoprotein (GP), making it an attractive target for antiviral drug development. The Ebola GP is a trimeric protein composed of two subunits per monomer: GP1, responsible for receptor binding, and GP2, which mediates fusion between the viral and host cell membranes. Initially synthesized as a precursor protein, the GP is cleaved by host proteases (furin, cathepsin) into its functional subunits, a process essential for its role in mediating viral entry. The GP facilitates the virus's attachment to the host cell surface, followed by conformational changes that enable membrane fusion, allowing the virus to enter the host cell (to the host endosomal Niemann-Pick C1 (NPC1) receptor or via direct membrane binding; Vaknin et al.; ACS Infect. Dis. 2024, 10, 5, 1590–1601).
Targeting the GP for drug development is advantageous due to its essential role in viral infection, its highly conserved structure among different Ebola virus strains, and the availability of specific binding cavities that can accommodate small-molecule inhibitors. Structural studies using techniques such as X-ray crystallography have identified these binding cavities and elucidated the GP's conformation in both its free and inhibited states. These insights enable the design of drugs that can specifically bind to and inhibit the GP by stabilizing it in its pre-fusion conformation or interfering with its cleavage, thereby preventing the necessary conformational changes for membrane fusion. We will employ high-resolution structures to conduct virtual screening experiments coupled to molecular dynamics simulations to ultimately identify potential GP inhibitors/modulators.
Promising compounds identified through these computational methods will hopefully undergo further validation using biochemical assays, pseudovirus entry assays, and structural analyses to confirm their inhibitory activity. Targeting the GP offers specificity, as it minimizes off-target effects on host cells and reduces the likelihood of resistance development. Moreover, due to the conserved nature of the GP, drugs targeting it could be effective against multiple Ebola virus strains and variants.
We hope that our computations will contribute to the fight against Ebola!
11 Jul 2024, 9:02:55 UTC
· Discuss
Vote for the next target!
Dear all, we have obtained results for different targets of SARS-CoV-2. The computations continue, and we want to ask your opinion on the next target. You can vote for one of them until February 5th, 2024.
- SARS-CoV-2 main protease (3CLpro) further studies
This is a crucial therapeutic target against SARS-CoV-2. 3CLpro (cysteine protease; EC 3.4.22.69) in particular is crucial for the cleavage of coronavirus polyproteins to form mature non-structural proteins that are themselves essential for viral replication mechanisms. We still need much more research on this target towards new more potent inhibitors.
- Ebolavirus glycoprotein (GP)
Ebola virus is a dangerous pathogen to humans and this target could be a perfect study case for PPI-type drug design scenario. The EBOV glycoprotein (GP) is the only virally expressed protein on the virion surface and is critical for attachment to host cells and catalysis of membrane fusion.
- Swine acute diarrhea syndrome coronavirus (SADS-CoV PLpro)
This target would enable us to study the design on multiple related viral targets. Swine acute diarrhea syndrome coronavirus (SADS-CoV), a newly emerging enteric coronavirus, is considered to be associated with swine acute diarrhea syndrome (SADS) which has caused significantly economic losses to the porcine industry. The pathogen indicates towards host-jump potential.
26 Jan 2024, 12:36:20 UTC
· Discuss
Project server maintenance 17th January 2024
Dear participants, tomorrow, 2024.01.17 due to server maintenance, the project will stopped for several hours.
Thank you for attention and project participation!
16 Jan 2024, 12:54:07 UTC
· Discuss
Project status: December 2023
Dear SiDock@home participants,
We've successfully achieved 21 milestones in our ongoing drug discovery initiative, and this strong, open, and community-supported drug discovery project is going on. Our research has been routine lately: virtual screening on the same library for a pleiade of corona-related targets. These efforts are however crucial to our research progression. Currently, we're in the process of drafting publications for two of our completed objectives (3CLpro and PLpro) and are setting the stage for upcoming drug targets (here we are also planning a pool where You the participants, will help us decide on the upcoming target work).
Last but not least, we thank everyone who has donated cryptocurrency or money. The donations sum up to 639 Euro and 12,140 Gridcoin now. We plan to use them for purchase of compounds and in-vitro screening. As always, we are grateful to all of you for your computational contributions and discussions!
We look forward to the future work on SiDock@home.
Merry Christmas to all and all the best to All!
With best wishes,
Natalia, Marko, Črtomir and hoarfrost
26 Dec 2023, 19:37:17 UTC
· Discuss
Changes in task scheduling and monitoring mechanism
Dear participants, at the start of the current search, we set a deadline for "CmDock long tasks" to 14 days. But the average time required to process a task is 40 hours. Also, we see many of uncompleted tasks which prevent completion of the set to which they belong, "produced" by the computers that get tasks and do not get in touch again. Due to these reasons, we plan:
1. To perform a gradual reduction of the deadline;
2. To implement a simple mechanism which finds computers that are lost in computing space (those who did not send requests to the server in the last m days), and mark their tasks as lost. For example, m may be 10, 8, 6 or 5.
This should not interfere with computers actually participating in the calculations, but can greatly enhance the logistics of results.
6 Nov 2023, 10:05:37 UTC
· Discuss
BOINC merchandise by the Science Commons Initiative
Dear participants,
There is a wonderful new beginning started by the Science Commons Initiative. They launched a web store selling BOINC-related merchandise. SiDock@home is already on board :)
We support this idea and hope it will help more people to know about BOINC and get interested in BOINC projects! It will also contribute to the development of BOINC as 50% of the profits will go to the BOINC Development Fund (100% in special cases). Another 50% of the profit will go to the project as a donation.
With best wishes,
team of SiDock@home
23 Feb 2023, 9:29:38 UTC
· Discuss
Target # 22: corona_RdRp_v2
Dear participants,
After doing initial studies on an established SARS-CoV-2 target RNA dependent RNA polymerase (RdRp), we are departing from protease studies and focus on RdRp along with additional NSP targets. Namely, RdRp (NSP12), is responsible for the transcription of viral genes and ultimately replication of the viral genome. The studied active site binds RNA and was previously studied in the context of remdesivir, galidesivir, molnupiravir and several other small molecules. Further reading below:
- Structure of replicating SARS-CoV-2 polymerase
- Identification of novel SARS-CoV-2 RNA dependent RNA polymerase (RdRp) inhibitors: From in silico screening to experimentally validated inhibitory activity
- RNA dependent RNA polymerase (RdRp) as a drug target for SARS-CoV2
- Ribavirin, Remdesivir, Sofosbuvir, Galidesivir, and Tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): A molecular docking study
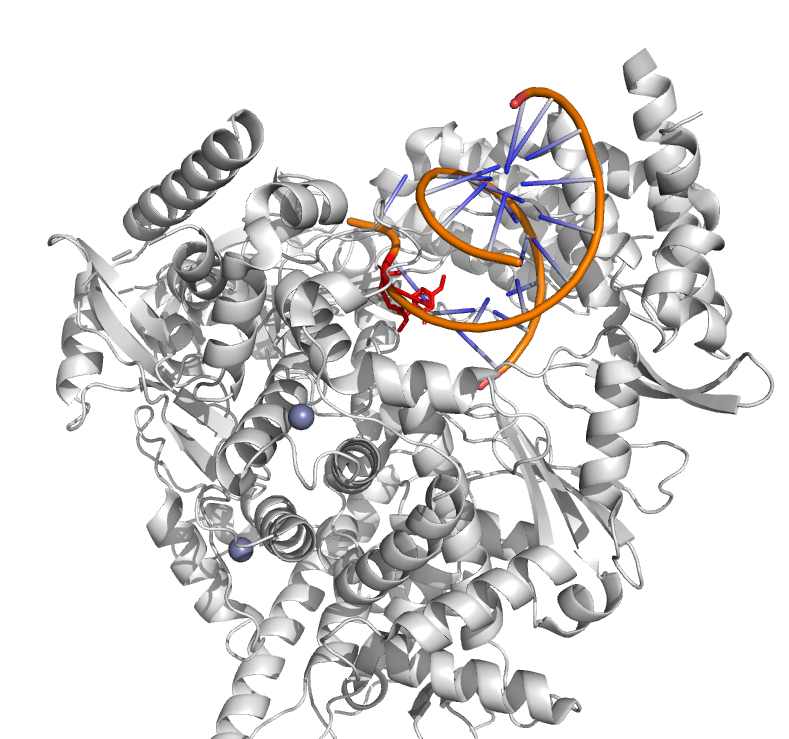
Figure 1: RdRp in white color along with RNA chain and N4-hydroxycytidine from Molnupiravir in the active site (red stick model).
With best wishes,
Natalia, Marko, Črtomir, hoarfrost.
15 Jan 2023, 17:45:23 UTC
· Discuss
СmDock "long" and "short" tasks applications
Dear participants!
We created new applications named as "CurieMarieDock 0.2.0 long tasks" and "CurieMarieDock 0.2.0 short tasks", both based on CmDock 0.2.0 release. Algorithm of both applications are identical, but for "short tasks" we created only ARM-version and we plan to generate a special, smaller in 5 times tasks that covers one of part of entire tasks set. We advice all owners of small single-board computers like Raspberry Pi, to switch to "short tasks" application.
In next 1 .. 2 days we continue to generate new tasks "Sprot_delta_v1" (extended run) for both application, but later we issue a sample tasks set for new target (# 22) and only for "long application" because all task of sample set will be large. Simultaneously, project will continue to send Sprot_delta_v1 tasks for "short" application. And then we will proceed to sent long and short tasks for appropriate applications.
Hope that this explanation makes situation is clear.
Thank you for participation and donation of CPU time!
13 Jan 2023, 23:35:04 UTC
· Discuss
Merry Christmas!
Dear participants,
We wish you a merry Christmas and a happy New Year!
With your help, we have performed a lot of computer modeling. We are going to continue, and believe the results will lead to advances in drug development! For now, we have a fresh new release of CmDock and will employ it starting from the next target. Here is an illustration of the modeling performed in the project, visualized with PyMOL. You can see a large molecule (this is the target, TMPRSS2 protein) with a molecule many times smaller. The small molecules are ligands, each of them can interact with the target in its own way, with its own binding energy.
The more precisely the ligand binds to the target, the higher the chance that it will be able to change the functioning of the large molecule. For example, the TMPRSS2 enzyme shown in the image is necessary for a chemical reaction allowing the coronavirus to enter the body cells and, accordingly, a substance that can suppress the normal operation of this enzyme can make it difficult or block the spread of the coronavirus in it. During the modeling performed on your computers, various ligands from the library are tested, and the best ones are selected to process on next stages of drug development.
All the best,
Natalia, Marko, hoarfrost, Črtomir
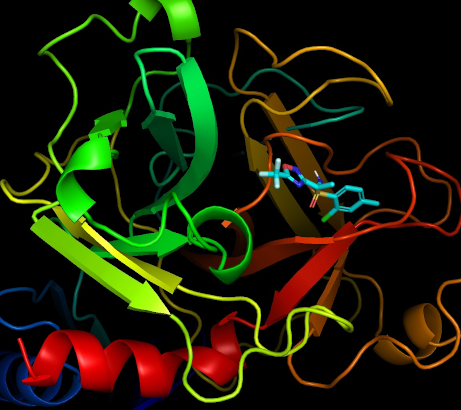
24 Dec 2022, 17:13:03 UTC
· Discuss
Release CmDock v 0.2.0
Dear SiDock@home community,
CmDock 0.2.0 is released:
-Cross-platform consistent random number generator (custom implementation of probability density functions); new switch -x defined, which turns on this feature.
-CI pipeline with regression testing
-logging in cmcavity
-cmcavity implementaiton of cavity volumes in InsightII (-I) or MRC 2014 (-M)
-Full protein surface mapping
-Pymol GUI plugin for Your demo/teaching/visualisation/research projects ;)
You can get it at:
https://gitlab.com/Jukic/cmdock/
Happy holidays to all!
, and all health!
Marko & CmDock team & SiDock@home team
14 Dec 2022, 16:02:58 UTC
· Discuss
BOINC Census 2022, be counted!
The BOINC Census is a yearly effort aimed at finding out who uses BOINC, what they like about it, and what changes they want to see in the future. Filling out the census is a great way to support this project, the wider BOINC community, and have your input heard for future bug bounties. The census is sponsored by the Science Commons Initiative, and the results will be published in an aggregate, anonymized fashion at the completion of the census. Please take 5 minutes to fill out the census at https://app.fillout.com/flow/esDGuRswWfus
17 Nov 2022, 13:24:51 UTC
· Discuss
Target 21: corona_TMPRSS2_v1
Dear Participants,
TMPRSS2 is a type II transmembrane protease with broad expression in epithelial cells of the respiratory and gastrointestinal human tracts. It is a cofactor in SARS-CoV-2 entry, and primes viral proteins. Namely, TMPRSS2 cleaves the viral Spro to expose the fusion peptide for cell entry. We are therefore examining TMPRSS2 as a promising strategy to block viral infection in a host-directed therapeutic and/or prophylactic manner. This work is also a a part of larger viral-host protease inhibition study campaign.
With best wishes,
Team SiDock@home
17 Nov 2022, 13:21:56 UTC
· Discuss
Temporarily work unavailabilities
Dear participants, after sending last tasks for target 20 (corona_DHODH_v1) we will start the experiment with one of already processed targets but with new parameters. Results for this tasks will be large and we will slow down the creation of new tasks for reducing load on network infrastructure. Often there will be no tasks.
Thank you for project support!
24 Oct 2022, 23:25:21 UTC
· Discuss
CmDock new release pending
Dear SiDock@home community,
Just a little update on the development of our CmD software. We were planning to release CmD 0.1.5.; however, we decided to wait a bit. Namely, we want to implement a robust testing (regression, unit) pipeline and lay the work for future software optimization. Therefore we would like to implement a cross-platform consistent pseudo-random number generation that would mean - write once, compile and test everywhere... We think this can be achieved relatively quickly.
On the other front, Mac builds are being tested and we will provide the community with Mac builds through homebrew platform. We will publish the news on future release here.
All the best,
Marko
22 Jul 2022, 15:33:02 UTC
· Discuss
Project status: May 2022
Dear all,
with your help, we have processed 15 targets already! It is a huge computational work, the results of which are being processed by our team and colleagues. We are working on two papers which will be available to the public as early as possible. We also continue computations for next targets. During March, April and May, project's performance was fluctuating around 250 Teraflops. We took this advantage to prioritize SARS-CoV-2 targets on top of the other research. At the end of spring 2022, we are 6426 active participants with 4493 computers.
With best wishes,
Natalia and the team of SiDock@home
31 May 2022, 19:44:33 UTC
· Discuss
Project status: March 2022
Dear SiDock@home community,
Here is an update on our work. This international scientific drug
discovery project is well on its way towards its first milestone of
experimental support for the docked compounds. We are working hard
towards data processing and scientific collaborations that can help us
take our proposed compounds further. In the near future, we will prepare
a scientific write-up for You to follow.
This international drug discovery project is a wonderful example of
community, citizen science, teamwork, and contributing to the field of
medicinal chemistry with the ultimate goal of contributing to the
wellbeing of us all. Our heart goes out to all!
We also thank You for the help and support that keeps us going.
Currently, our servers reside in Russia and Slovenia. This enables us
easier task allocation and backup of the results and we hope to add a
third location with additional hardware in the future. Due to the
community request, we also plan to add official Mac and ARM64 support
for the underlying CmDock software.
Moreover, we are continuing with a new target, still from the SARS-CoV-2
pathogen, namely nsp14. It consists of the N-terminal ExoN domain
involved in proofreading and the C-terminal guanine N7 methyl
transferase (N7-MTase) domain that functions in mRNA capping and on the
latter we are focusing on now.
After this target is complete, we are turning our attention towards one
potential therapeutic target of the Ebola viral pathogen to return back
to SARS-CoV-2 after that target will be complete.
We are committed to this drug discovery project and the online
community, hope to expand and we look forward to working and discovering
new molecules and compound mechanisms of action in the future.
All health, luck, and peace to all!
Marko Jukic
14 Mar 2022, 13:58:39 UTC
· Discuss
Technical maintenance on March 14th, 9:00-10:00 UTC
Dear participants,
we plan an hour-long break tomorrow, 9:00-10:00 UTC, for technical works.
All the best,
Team SiDock@home
13 Mar 2022, 16:18:05 UTC
· Discuss
Project maintenance on February 26
Dear participants, new portion of project maintenance is scheduled on Ferbruary 26th, 10:00 UTC, for ~1 hour.
Thank you for participation!
26 Feb 2022, 9:02:37 UTC
· Discuss
The project is online!
Dear participants!
The work by upgrade is completed. Project database, website and request processing migrated on pair of mirrored SSD, workunit files (as previously) and uploaded results placed on mirrored HDD.
Thank you for attention and participation!
17 Feb 2022, 13:54:21 UTC
· Discuss
Technical maintenance on February 15th
Hello! Additional server maintenance planned on February 15th, for several hours. Approx. 08:00-20:00 UTC.
15 Feb 2022, 7:01:45 UTC
· Discuss
Technical maintenance on February 14th
Dear all,
On February 14th, for about 08:00-20:00 UTC, the project server will be shut down for maintenance. We will upgrade hard drives to support larger loads.
All the best,
Team SiDock@home
11 Feb 2022, 13:50:35 UTC
· Discuss
Target 11 - Sprot_delta_v1
Dear Participants,
We continue diligently with the calculations. Now we are breaking through a series of protease studies to explore a new target, the RBD (receptor-binding domain) Sprot domain surface interacting with ACE2. This is the much-discussed spike protein that also contributed to the naming of coronavirus. We focus here on Sprot from the delta variant (B.1.617.2), specifically a Cryo-EM structure of Sprot with an RBD in up conformation. In comparative variant studies, we will try to determine if there is a specific discrimination upon binding of small ligands. This may help us better understand this PPI surface in further studies and even develop a small molecule probes to study these interactions.
Thank You all and we look forward to future studies together!
With best wishes,
Team SiDock@home
10 Feb 2022, 13:45:49 UTC
· Discuss
SiDock@home gets supported by the Charity Event 2022 (15 - 30 January)
Dear folks!
Charity Event team invites you to take part in the Charity Event 2022 addressing SiDock@home. You can read more about this event in thread on project forum and general forum of all Charity Events.
Thank you for attention to the project and participation!
6 Jan 2022, 17:00:32 UTC
· Discuss
Merry Christmas!
Dear participants,
Merry Christmas and a happy New Year to all of you! We wish you lots of health, love and joy.
With Your help, we have done an enormous research work, and we are preparing to do even more and take this drug discovery project even further. We will be tackling future key problems in drug design and are working hard to overcome all obstacles of financing (with your help it is easier… thank You all!!!) and wet lab support!
Currently, we are finishing the 5th target and also returning to 3CLpro allosteric sites towards further in depth research.
All the best! From Natalia, Marko, Črtomir and hoarfrost
26 Dec 2021, 13:20:13 UTC
· Discuss
A poll about distributed computing projects
Dear all,
there is a poll created by students who investigate volunteer computing. The questions aim to explore the motivation of participants. If you wish and have a minute, please express your opinion at https://forms.gle/tcLYdtoeVfMMREPE7. The results will be analyzed and presented to the public. Thank you!
With best wishes,
Team of SiDock@home
24 Nov 2021, 11:51:31 UTC
· Discuss
Release CmDock v 0.1.4
Dear community. New release of CmDock was prepared. The v0.1.4 of CmDock incorporates fixed checkpointing and progress bar patch along with support for compressed input and output via cmzip. New CmDock release also includes an Updated SDF-Tools submodule that should help the userbase manipulate input/output files with support for large molecular libraries.
Along with the new release, BOINC client is up to date also. The team is working hard and we are committed to support CmDock in the future. Thank You!
Marko & Natalia
21 Nov 2021, 17:10:00 UTC
· Discuss
Ninth and tenth targets
Dear participants,
We have a new target which resides at a novel allosteric site on SARS-CoV-2 main protease. The site was identified by MixMD method [1] with collaboration at College of Pharmacy, University of Michigan. Target preparation was helped by Jelena Tošović (Slovenia).
The two upcoming targets and results will complement the allosteric sites identified with X-ray screening by Sebastian Günther et al. [2]. The targets have good chances for wet-lab testing as soon as possible and further experimental support.
(We will again temporarily pause processing of Target 5)
With best wishes,
Team SiDock@home
[1] Phani Ghanakota and Heather A. Carlson. Moving Beyond Active-Site Detection: MixMD Applied to Allosteric Systems. The Journal of Physical Chemistry B 2016 120 (33), 8685-8695. DOI: 10.1021/acs.jpcb.6b03515
[2] Sebastian Günther et al. X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease. Science 2021 372 (6542), 642-646. DOI: 10.1126/science.abf7945
13 Nov 2021, 9:39:16 UTC
· Discuss
November competition
Dear all,
we are glad to announce a November competition! The symbol of the competition is tryptophan. Just like any amino acid, tryptophan serves as a building block in protein biosynthesis. It is also a source to synthesize serotonin, melatonin, and vitamin B3. And just like many vital processes in our bodies, tryptophan biochemistry is affected by SARS-CoV-2 infection ([1-3] etc.).
1st place: 
2nd place: 
3rd place: 
[1] Thomas T, Stefanoni D, Reisz JA, et al. COVID-19 infection alters kynurenine and fatty acid metabolism, correlating with IL-6 levels and renal status. JCI Insight. 2020;5(14):e140327. Published 2020 Jul 23. doi:10.1172/jci.insight.140327
[2] Eroğlu İ, Eroğlu BÇ, Güven GS. Altered tryptophan absorption and metabolism could underlie long-term symptoms in survivors of coronavirus disease 2019 (COVID-19). Nutrition. 2021;90:111308. doi:10.1016/j.nut.2021.111308
[3] Vyavahare S, Kumar S, Cantu N, et al. Tryptophan-kynurenine pathway in COVID-19-dependent musculoskeletal pathology: a minireview. Mediators of Inflammation. 2021; Article ID 2911578, 6 pages, 2021. doi:10.1155/2021/2911578
28 Oct 2021, 19:08:55 UTC
· Discuss
BOINC client 7.16.20 solves the certificate problem for Windows
Dear all,
If you are running BOINC on a Windows computer, please update to a new version (7.16.20) of the BOINC client. The previous version stopped working recently because of an expired SSL file that prevented it from communicating with several BOINC projects, with the BOINC server, and with Science United. You can download the new version here: https://boinc.berkeley.edu/download.php
With best wishes,
Team of SiDock@home
20 Oct 2021, 8:03:32 UTC
· Discuss
Certificates and project communicating
Dear folks!
If BOINC on your computer experiencing a problem during project communicating, try to replace file ca-bundle.crt in programs directory of BOINC (C:\Program Files\BOINC\ca-bundle.crt, for example) by this file: https://srbase.my-firewall.org/sr5/download/ca-bundle.crt and restart whole BOINC processes or reboot computer.
Another threads on forum:
Peer certificate cannot be authenticated with given CA certificates
Wus not uploading
Transient Error
Communication deferred
100% block still crunching
Thank you for participation!
4 Oct 2021, 18:55:28 UTC
· Discuss
4000 users with credit
Dear SiDockers,
new breakthrough of the project, we will wellcome 4000th user with the credit, very soon.
;-) Crtomir
21 Sep 2021, 15:17:51 UTC
· Discuss
SiDock@home September Sailing
Dear participants!
Just now, your computers are hitting three protein targets at once. Due to the low runtimes and (mainly) the friendly contributions of participants and their teams, the data is being processed fast, and we express our gratitude to all of you.
We announce a new competition between teams, SiDock@home September Sailing. Top winners will get the badges:
1st place:
2nd place:
3rd place:
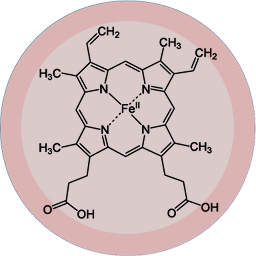
Heme B, the well-known ligand in the complex of Hemoglobin, helps to carry oxygen in our bloodstream. As we know today, it has emerged from just two mutations that occurred in genome of one of predecessors of modern vertebrates more than 400 million years ago [1,2].
[1] https://phys.org/news/2020-05-reveal-complex-hemoglobin-resurrecting-ancient.html
[2] Pillai, A.S., Chandler, S.A., Liu, Y. et al. Origin of complexity in haemoglobin evolution. Nature 581, 480–485 (2020).
12 Sep 2021, 8:19:15 UTC
· Discuss
Targets six, seven, eight
Dear participants,
we have prepared three new targets for you! They all base on 3CLpro protein, a prime target for antiviral drug discovery because of its central involvement in virus replication. Another reason to put them high on the list is that we have a good possibility to test interesting results in a lab. The targets 6, 7 and 8 are X-ray identified allosteric sites on 3Clpro. We will make a limited number of workunits to ensure everything works fine, and then launch full-scale screening on the new targets. We will temporarily pause processing of Target 5 in favour of the new ones.
With best wishes,
Team SiDock@home
28 Aug 2021, 16:18:49 UTC
· Discuss
Project status: July 2021
Dear all,
Our research is going on! Collaborators just started biological evaluation of 30 chemical compounds selected in SiDock@home for targets 3CLpro and PLpro. Biological evaluation in vitro works with the most prospective results that your computers have selected in silico. The process is iterative and will involve hundreds of experiments.
With your help, we have already processed 4 targets and 43% of the fifth one. Further workunits will be created basing on the biological evaluation and the latest scientific knowledge about the targets.
Many thanks and best wishes,
Team of SiDock@home
9 Jul 2021, 18:21:50 UTC
· Discuss
Donations to SiDock@home
Drug development is a time-consuming and costly process. With help of your computers, our team is able to speed up discovery of prospective candidates for in vitro tests. At the same time, the volume of biological evaluation is limited by funding. We are in constant search for additional sources, and, since SiDock@home allows the participants to earn Gridcoins, we find it a good idea to accept voluntary donations. If you feel like donating to the project, please use one of the listed options.
9 Jul 2021, 18:18:53 UTC
· Discuss
SiDock@home is whitelisted in Gridcoin
Hello everyone,
since today, SiDock@home is whitelisted in Gridcoin. It means that you can receive the Gridcoin cryptocurrency for contributing to SiDock@home. For instructions, please refer to the official website of Gridcoin. For a description how Gridcoin works, refer to their whitepaper. This technology is yet new to our team, but we will do our best to understand it and help you to use it.
28 Jun 2021, 12:23:44 UTC
· Discuss
SiDock@home is going to join Gridcoin
Hello everyone,
we want to be whitelisted in Gridcoin (anytime), so you will be able to receive this cryptocurrency for scientific computing. We believe that Gridсoin is ethical. This is also a chance to increase awareness of the project.
6 Jun 2021, 19:00:14 UTC
· Discuss
Release CmDock v 0.1.3
Dear participants!
The following changes will be transparent, but if you are interested in technical details, then here they are. A new application cmdock-boinc-zcp has been released, including the new CmDock v0.1.3 release with (z)ipped input, (c)heckpoints and enhanced (p)rogress bar. In addition, a new docking experiment will begin under a new protocol.
Thanks to the CmDock team and pschoefer & walli for the release!
28 May 2021, 11:07:44 UTC
· Discuss
Release CmDock v 0.1.2
Hello everyone,
We are glad to announce the new app version with checkpoints, CmDock v 0.1.2 (cmdock-boinc-zip 2.0 in the BOINC project). Currently it is implemented for Windows 64bit, Linux 64bit and Raspberry Pi (for RPi, please check this thread if you need instructions on configuring the client).
The new release has been tested, but not on every existing computer. In case of any problems, please report.
Thank you all for participation!
20 May 2021, 17:00:11 UTC
· Discuss
Half a year, the BOINC Workshop and a move to the new server
Hello all!
Today we celebrate six months of SiDock@home. Thank you for lots of support! Let us summarize the current state:
Science. We are post-processing the obtained results and simultaneously performing virtual screening for the 5th target. With results obtained for several targets, we focus on viral proteases and are in the phase of getting physical samples of compounds for wet-lab testing. We will present the project at the BOINC Workshop on April 28. The event starts at 09:00 PDT / 12:00 EDT / 18:00 CEST / 19:00 EEST, our talk is at +00:40.
Society. The BOINC Pentathlon team selected SiDock@home as this year's Marathon project. The Marathon discipline is announced on 30 April and runs for two weeks from 05 through 18 May.
Technicals. Before the BOINC Pentathlon, we plan to move to a new physical server. The URL will stay the same. But we stop generating tasks and the project will be unavailable for 1-2 days. Also, we are implementing checkpoints as soon as possible.
Best wishes,
SiDock@home team
23 Apr 2021, 10:19:03 UTC
· Discuss
BoincWorkshop 2
From chat in BoincWorksop Day 2
"I'm seeing far fewer people interested in running BOINC and many are giving up due to too few "legitimate" projects continuing their research - too many projects have used volunteers and then stopped."
"So, we need more "interesting" projects so people can crunch them...far more people got involved with SiDock, Rosetta and WCG when they launched Covid-19 research tasks..".
Our community is on the right way ;-)
Crtomir
21 Apr 2021, 19:10:17 UTC
· Discuss
Upcoming conference: ANTYCOVID-21 (online)
Dear all,
on June 28-29 2021, there will be a scientific conference "ANTYCOVID-21: Computer science protecting human society against epidemics", in an online format, hosted in Poland. We aim to present a talk about SiDock@home and will be glad to see all of you as listeners. We also encourage you to submit own works if you participate in a covid-related research!
Conference leaflet
Dates:
Paper submission 15 April 30 May (extended)
Notification of paper acceptance: 15 May
Final paper submission and registration: 30 May
Conference 28-29 June 2020
Submissions:
The papers should be submitted via Easychair
The submission may include long papers limited to 12 pages in Springer LaTex llncs format or short papers (up to 4 pages)
All questions about submissions may be emailed to antycovid21 at easychair.org
2 Apr 2021, 11:27:23 UTC
· Discuss
Fifth Target is SARS-CoV-2 EProtein
Our fifth target is the E protein. We are emotionally attached to this protein because we explored its druggability in the early days of our project in the context of Covid.
At only 75 amino acids long, the envelope protein (E) is the smallest of the four structural proteins that make up the SARS-CoV-2 virus particle, and it is essential for the virus to infect cells. Data from other coronaviruses led researchers to suspect that groups of five E proteins form a pore that spans the lipid bilayer membrane of the virus. However, there was no direct evidence because the structural characterization of membrane-spanning proteins is difficult with the most commonly used techniques - X-ray crystallography and cryo-electron microscopy. Mei Hong's group at the Massachusetts Institute of Technology instead used nuclear magnetic resonance spectroscopy to solve the structure of the E protein and confirm pore formation (Nat. Struct. Mol. Biol. 2020, DOI: 10.1038/s41594-020-00536-8). The group also investigated how two drugs, amantadine, and hexamethylene amiloride, can bind to and block the pore. Although these drugs only bind weakly to the pore, the researchers say the new structural information could help in the development of drugs that target the virus.
In our HTVS, we will screen 1 billion of the small molecules to the pore of the E protein pentamer. We hope to find some nice molecules that can block the pore ;-)
19 Mar 2021, 21:50:22 UTC
· Discuss
SiDock@home becomes a general drug discovery project
Hello everyone!
We are happy to announce that SiDock@home will continue as a general drug discovery project, not limited to coronavirus only! This means that the project will continue for a long time. We aim to develop and incorporate novel methods of drug design and perform drug discovery for different diseases.
Nevertheless, the first mission on fighting the coronavirus continues! There are many results to process and more targets to explore within what started from COVID.SI project.
With best wishes,
Natalia and the team.
27 Feb 2021, 14:06:38 UTC
· Discuss
Fourth target (corona_RdRp_v1)
We are glad to announce the fourth target, corona_RdRp_v1. This molecule is frequently used in research studies as a potential target to inhibit viral replication. Specifically, it is SARS-CoV-2 RNA-dependent RNA polymerase (RdRp in short), an enzyme which coronavirus SARS-CoV-2 uses for the replication of its genome and the transcription of its genes.
8 Feb 2021, 8:17:30 UTC
· Discuss
Release CMDock v 0.1.1
Official release of CurieDock (CMDock v 0.1.1) for windows, linux and arm (raspberry pi):
https://gitlab.com/Jukic/cmdock/-/releases
5 Feb 2021, 19:10:12 UTC
· Discuss
Third target (corona_PLpro_v2)
Hello everyone!
We are glad to announce work on the third target: PLpro_v2, the same protein with another catalytic active site.
We resume task generation with a new validator. It was implemented to automate result check which caused some delays before.
Thank you all for supporting the project!
29 Jan 2021, 10:21:40 UTC
· Discuss
Temporary task shortage
Hello everyone! There is a temporary task shortage due to finishing the 2nd target, moving to the 3rd one and improving the docking protocol. Thank you for your patience!
21 Jan 2021, 15:30:02 UTC
· Discuss
Milestones of January
Hello everyone!
The beginning of the year was productive: 1000 active users and 4000 computers!
With your help, we are finishing the 2nd target, working towards next targets and prospective testing in a wet-lab.
Thanks to everyone and good health.
Hoarfrost and Natalia.
18 Jan 2021, 6:24:36 UTC
· Discuss
Second target (corona_PLpro_v1)
Hello everyone! With the beginning of the year, we started processing of the second target, PL Pro. Many new participants joined, so almost half of the library has been docked. Excellent work! Thank you for contributing.
11 Jan 2021, 12:10:19 UTC
· Discuss
First target (3CLpro_v3) completion and outage of work
Hi folks!
Today we generated the last set of tasks for first target - "corona 3CL pro v3" protein. Also we generated additional set of tasks named like "corona_3CLpro_v3_ext_nb3di_0000493_2_replica". This is replica of tasks that we crunch previously and they need for test purpose - for estimate of possible methods of additional validation of results. In next few days project run out of tasks, but next target and tasks is coming soon!
Thank you for participation and Happy New Year! 2021 is coming!
30 Dec 2020, 22:46:02 UTC
· Discuss
CmDock source code
Hi everyone,
here is the CmDock source code repository: https://gitlab.com/Jukic/cmdock.
This is the main application that we use in the project. Now you can try and build it for your own platforms!
We have many plans and invite C++ devs, especially ones with CUDA and parallel computing knowledge to join our project.
If you would like to join, please contact Marko: jukic (dot) marko (at) gmail (dot) com.
With best wishes,
Natalia
29 Dec 2020, 8:48:13 UTC
· Discuss
Merry Christmas!
Merry Christmas to everyone who celebrates it by the Gregorian calendar! Let it bring joy and love!
We hope you will like the new badges for the total credit. New tasks will be soon. We work at making workflow more stable.
With best wishes, Natalia and hoarfrost.
25 Dec 2020, 7:15:06 UTC
· Discuss
SiDock@home
This is the production project, a followup to the testing we have been doing. You are welcome to join on our scientific route!
18 Dec 2020, 12:55:33 UTC
· Discuss
Transition to production project
Hi folks!
During the next few days we plan to transform the current test project into production. Today we generate last bunch of workunits and after its completion test project will go to shutdown. Production project will have another URL and we will publish it. Also we plan to move statistics of teams and participants, as well as accounts information from test to production project.
All of you are highly welcome to stay with us on the production project!
10 Dec 2020, 13:42:35 UTC
· Discuss
2020-12-04: progress report
Hello everyone!
Here is an overview of the work done so far.
Firstly, we have docked 48% of the ligands database to the first target, 3CL Pro. 480 million small molecules were tested in silico. This is a great job! Now we move on to cover 100% of the database.
Virtual screening is a complex procedure that includes post-processing of results, some of them redundant. So the next important step will be processing of the results and studying the chemical subspace they represent. It is important to understand how are the docking scores divided in the dataset. Then our team will develop more efficient filtering rules.
Secondly, the next target for docking is being prepared. Overall, there are 59 targets related to SARS-CoV-2 to investigate.
Thirdly, the new docking software CurieMarieDock has been developed as a fork of RxDock. It is optimised and runs faster. Windows, MacOS and Linux versions are released. ARM version is being prepared.
It is fascinating to work with all of you! Thank you for participating! Have a nice weekend.
With best wishes,
Natalia
4 Dec 2020, 18:18:08 UTC
· Discuss
New application: CurieMarieDock
Hello everyone!
We are glad to announce the new application CurieMarieDock (cmdock). Currently for Windows 64bit and MacOS 64bit, and more platforms will follow. If you observe errors or undesirable effects in it, please report.
With this application, there is no need for installing MS Visual C++ Redistributable for Visual Studio.
CurieMarieDock is a molecular docking application developed as a fork of RxDock by the authors of the basic COVID.SI project: Marko Jukić, Nejc Ilc, Davor Sluga, Črtomir Podlipnik, and Gašper Tomšič.
Thank you everyone for participating.
2 Dec 2020, 14:25:49 UTC
· Discuss
App version 12.2 (Linux 64bit)
Hi all! Please note release of a new Linux version 12.2, the tasks of which the BOINC client should now suspend normally.
Technically, there are two fixes:
1) Wrapper now launches docking application directly, without intermediate script.
2) Wrapper now sends SIGSTOP signal instead of SIGTSTP when suspending a task.
27 Nov 2020, 15:24:02 UTC
· Discuss
App version 12.2 (Windows 64bit)
Hi all! Please note release of a new Windows version 12.2, the tasks of which the BOINC client should now suspend normally. Technically, the wrapper now launches docking application directly, without intermediate .bat script.
23 Nov 2020, 15:07:00 UTC
· Discuss
App version 12.1 (Windows 64bit)
Please note we released a new app version 12.1 (Windows 64bit). Technically, we deleted the script of calculating task progress which caused many errors. Still, the BOINC client may display task progress using its own algorithms based on the history.
17 Nov 2020, 11:19:46 UTC
· Discuss
Disabling task generation for Windows
Dear folks, we temporary disable tasks generation for Windows due to high rate of errors on Windows machines. This application needs more work.
16 Nov 2020, 11:40:05 UTC
· Discuss
App version 12.0 (Windows 64bit)
Please note we released a new app version 12.0 (Windows 64bit) which should resolve errors on Windows 7.
16 Nov 2020, 10:53:54 UTC
· Discuss
News is available as an RSS feed
©2026 SiDock@home Team